Cell-weighing method could help doctors choose cancer drugs
Doctors have many drugs available to treat multiple myeloma, a type of blood cancer. However, there is no way to predict, by genetic markers or other means, how a patient will respond to a particular drug. This can lead to months of treatment with a drug that isn’t working.
Researchers at MIT have now shown that they can use a new type of measurement to predict how drugs will affect cancer cells taken from multiple-myeloma patients. Furthermore, they showed that their predictions correlated with how those patients actually fared when treated with those drugs.
This type of testing could help doctors predict drug responses based on measurements of cancer cell growth rates after drug exposure, says Scott Manalis, the Andrew and Erna Viterbi Professor in the MIT departments of Biological Engineering and Mechanical Engineering and a member of MIT’s Koch Institute for Integrative Cancer Research.
“For infectious diseases, antibiotic susceptibility testing based on cell proliferation has been extremely effective for many decades,” Manalis says. “Unlike bacteria, analogous tests for tumor cells have been challenging, in part because the cells don’t always proliferate upon removal from the patient. The measurement we developed doesn’t require proliferation.”
Manalis is the senior author of the study, which appears in the Nov. 20 issue of Nature Communications. The paper’s lead authors are Mark Stevens, a visiting scientist at the Koch Institute and research scientist at Dana-Farber Cancer Institute, and Arif Cetin, a former MIT postdoc.

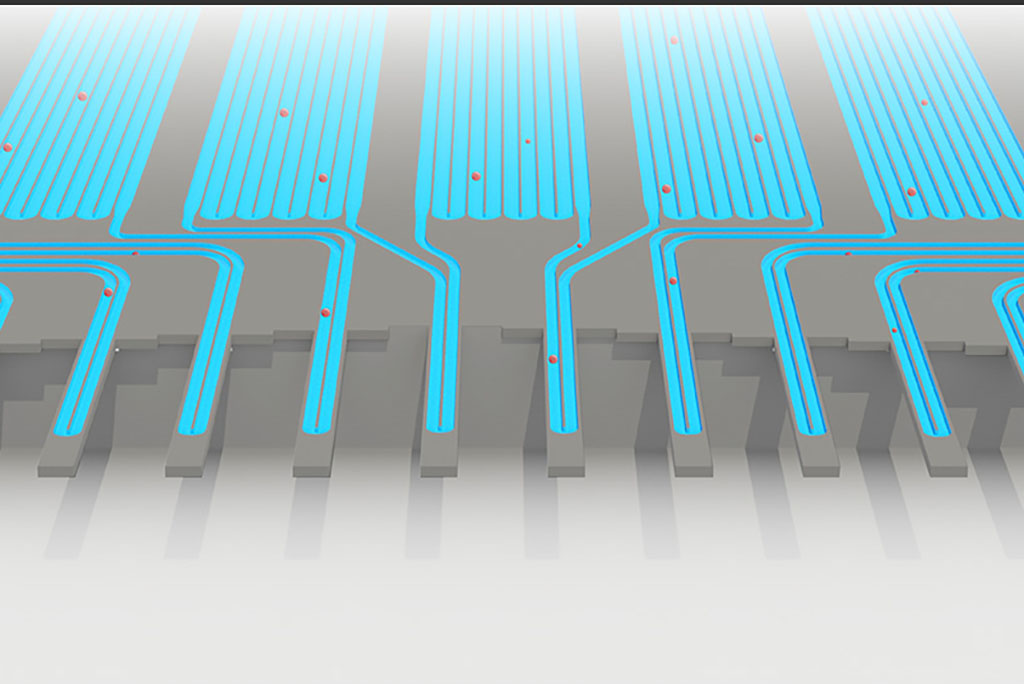
An animation of how a serial suspended micro channel resonator functions with cells flowing through the device. Cells pass over 10 cantilever mass sensors to have their mass weighed over a 15 minute interval, with measurements spaced by delay channels. Multiple measurements allow the researchers to capture changes in mass over time, a metric called mass accumulation rate (MAR).
Predicting response
The researchers’ new strategy is based on technology that Manalis and others in his lab have developed over the past several years to weigh cells. Their device, known as a suspended microchannel resonator (SMR), can measure cell masses 10 to 100 times more accurately than any other technique, allowing the researchers to precisely calculate growth rates of single cells over short periods of time.
The latest version of the device, which can measure 50 to 100 cells per hour, consists of a series of SMR sensors that weigh cells as they flow through tiny channels. Over a 20-minute period, each cell is weighed 10 times, which is enough to get an accurate MAR measurement.
A few years ago, Manalis and colleagues set out to adapt this technique to predict how cancer drugs affect tumor cell growth. They showed last year that the mass accumulation rate (MAR), a measurement of the rate at which the cells gain mass, can reveal drug susceptibility. A decrease in MAR following drug treatment means the cells are sensitive to the drug, but if they are resistant, there is no change in MAR.
In the new study, the researchers teamed up with Nikhil Munshi at Dana-Farber Cancer Institute to test a variety of drugs on tumor cells from multiple-myeloma patients. They then compared the results to what happened when the patients were treated with those drugs. For each patient, they tracked the cells’ response to three different drugs, plus several combinations of those drugs. They found that in all nine cases, their data matched the outcomes seen in patients, as measured by clinical protein biomarkers found in the bloodstream, which are used by doctors to determine whether a drug is killing the tumor cells.
“When the clinical biomarkers showed that the patients should be sensitive to a drug, we also saw sensitivity by our measurement. Whereas in cases where the patients were resistant, we saw that in the clinical biomarkers as well as our measurement,” Stevens says.
Personalized medicine
One of the difficulties in treating multiple myeloma is choosing among the many drugs available. Patients usually respond well to the first round of treatment but eventually relapse, at which point doctors must choose another drug. However, there is no way to predict which drug would be best for that particular patient at that time.
In one scenario, the researchers envision that their sensor would be used at the time of disease relapse, when the tumor may have developed resistance to specific therapies.
“At this time of relapse, we would take a bone marrow biopsy from a patient, and we would test each therapy individually or in combinations that are typically used in the clinic. At that point we’d be able to inform the clinician as to which therapy or combinations of therapies this patient seems to be most sensitive or most resistant to,” Stevens says.
The new test holds “great promise” to screen myeloma cells for drug susceptibility, says Kenneth Anderson, a professor of medicine at Harvard Medical School and Dana-Farber Cancer Institute, who was not involved in the research.
“This assay may fast-forward personalized medical care and the choice of effective therapies for myeloma both at diagnosis and at relapse,” Anderson says. “It may also be useful to profile susceptibility of minimal residual disease in order to further inform therapy and improve patient outcome.”
Bone marrow biopsies often produce limited numbers of tumor cells to test — as few as 50,000 tumor cells in this study — but for this technique that is enough to test many different drugs and drug combinations. The MIT researchers have started a company to begin a larger clinical study for validating this approach, and they plan to investigate the possibility of using this technology for other types of cancer.
The research was funded by the National Institutes of Health, the Koch Institute’s Cancer Center Support (core) Grant from the National Cancer Institute, the Department of Veterans Affairs, a Koch Institute Quinquennial Cancer Research Fellowship, and the Bridge Project of the Koch Institute and Dana-Farber/Harvard Cancer Center.